Abstract
Introduction: AML is a heterogeneous disease with a wide array of common genetic aberrations. Traditional classification of AML leverages both classical cytogenetics and mutational profiling to stratify patients into four distinct risk groups (ELN). However, tumor gene expression profiles can play an important role in response to therapy, and are potentially useful for unravelling the heterogeneity of AML. In this study, we hypothesized that clinical outcomes and variable responses to therapeutic modalities in AML may be driven by patterns of gene expression, and sought to identify clinically actionable molecular subtypes using the available RNAseq data from the BEAT AML functional genomics study.
Methods: Unsupervised machine learning approach based on consensus non-negative matrix factorization (cNMF) was applied to VOOM normalized BEAT-AML RNAseq data from patient samples with ≥50% blasts (N=389) to identify transcriptomic-based molecular subtypes. The subtypes were then compared to the genomic based subtypes for their association with clinical outcome (log-rank test) and ex-vivo drug sensitivity (Kruskal Wallis test). Subtypes were also biologically characterized by gene signature scoring using well curated pathway signatures (GSVA analysis using Hallmark pathways), cell type enrichment (xCell enrichment) and AML differentiation state (scRNAseq signature based on Van Galen et. al). Finally, a random forest classifier was defined based on samples from BEAT AML to predict the NMF subtypes in an independent data set (TCGA AML cohort).
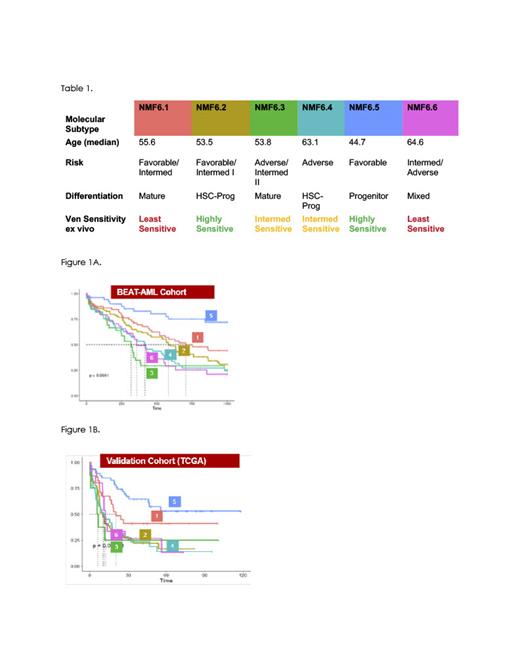
Results: Our cNMF based analysis identified six clusters of patients based on the 5,060 (top 10%) most variable genes. These novel subtypes were strongly prognostic (Figure 1A, log rank p=2.79e-08), and were independent of ELN genomic based subtypes (anova p=4.45e-07). Comparison to other genomic based classification is ongoing. The prognostic value of the transcriptomic subtypes was further validated by predicting the subtypes in an independent cohort (TCGA LAML, N=200). We observed a significant association with outcome (Figure 1B, p=0.00013), with clusters 5 and 1 showing markedly better prognosis, similar to BEATAML. These subtypes also displayed unique biological profiles, including significant association with scRNAseq-derived AML differentiation state cell types, Hallmark pathways and cellularity signatures. Notably, clusters 1 and 3 showed a mature phenotype, while clusters 2, 4, and 5 were more progenitor-like (table 1).
Importantly, the transcriptomic subtypes were highly predictive of ex-vivo drug sensitivity, with sensitivity to 70 compounds significantly associated with cNMF subtype (Kruskal Wallis p>0.01), compared with 4 in the ELN subtypes.Of the tested molecules, single agent Venetoclax was the most strongly associated with subtype (p=1.7e-13); two subtypes were strongly resistant (median IC50 of 10uM) and four were sensitive, with IC50s in the sub-micromolar range (Table 1). No association was seen between the ELN subtypes and venetoclax sensitivity (p=.35).
Conclusions: Unsupervised machine learning-based clustering analysis of transcriptomic data identified six novel subtypes which are similarly prognostic as the ELN genomic based subtype and provide a novel avenue for identifying clinically actionable subsets of AML.
Hamidi: Genentech: Current Employment, Current equity holder in publicly-traded company. Bolen: Genentech: Current Employment; F. Hoffmann-La Roche: Current equity holder in publicly-traded company. Lasater: Genentech: Current Employment, Current equity holder in publicly-traded company. Dunshee: Genentech/Roche: Current Employment, Current equity holder in publicly-traded company. Punnoose: Genentech: Current Employment, Current equity holder in publicly-traded company. Dail: Genentech/Roche: Current Employment, Current equity holder in publicly-traded company.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal